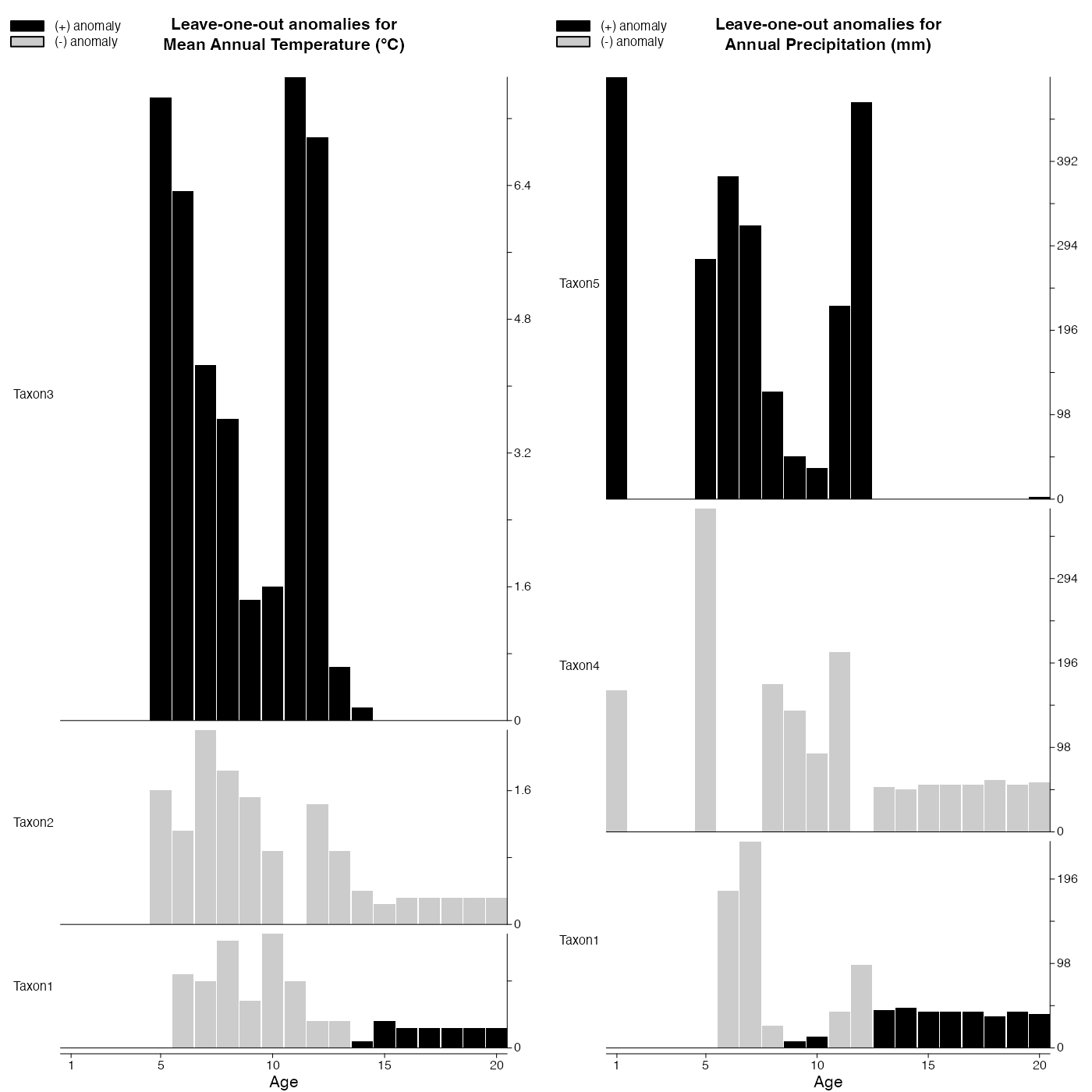
Repeat the reconstructions by removing one taxon at a time.
loo(x, climate = x$parameters$climate, verbose = TRUE)Arguments
- x
a
crestObjproduced by thecrest.reconstructorcrestfunctions.- climate
A vector of the climate variables to extract. See
accClimateVariablesfor the list of accepted values.- verbose
A boolean to print non-essential comments on the terminal (default
TRUE).
Value
A crestObj object containing the reconstructions and
all the associated data.
Examples
if (FALSE) { # \dontrun{
data(crest_ex)
data(crest_ex_pse)
data(crest_ex_selection)
reconstr <- crest(
df = crest_ex, pse = crest_ex_pse, taxaType = 0,
climate = c("bio1", "bio12"), bin_width = c(2, 20),
shape = c("normal", "lognormal"),
selectedTaxa = crest_ex_selection, dbname = "crest_example"
)
reconstr <- loo(reconstr)
} # }
## example using pre-saved reconstruction obtained with the previous command.
data(reconstr)
lapply(reconstr$reconstructions$bio12$loo, head)
#> $Taxon1
#> optima mean
#> [1,] 0.0000 0.0000
#> [2,] 0.0000 0.0000
#> [3,] 0.0000 0.0000
#> [4,] 0.0000 0.0000
#> [5,] 0.0000 0.0000
#> [6,] -182.3647 -197.7712
#>
#> $Taxon2
#> [1] NA
#>
#> $Taxon3
#> [1] NA
#>
#> $Taxon4
#> optima mean
#> [1,] -164.1283 -183.5160
#> [2,] 0.0000 0.0000
#> [3,] 0.0000 0.0000
#> [4,] 0.0000 0.0000
#> [5,] -375.1503 -373.2838
#> [6,] 0.0000 0.0000
#>
#> $Taxon5
#> optima mean
#> [1,] 489.7796 408.2192
#> [2,] 0.0000 0.0000
#> [3,] 0.0000 0.0000
#> [4,] 0.0000 0.0000
#> [5,] 278.7575 218.4514
#> [6,] 375.1503 310.0513
#>
plot_loo(reconstr)